| Browse | Text search | Protein search | Domain search | Download | User manual | Related links | Citation & Contact |
The Outer membrane porin (OprD) family [Function: Specific diffusion channels] Seed alignment | Full alignment | Pfam page | Pfam Wiki page | TC-DB page | ||||
The OprD family includes a number of porins in Pseudomonas species and other Gram-negative bacteria that appear to exhibit a variety of substrate specificities. OprD facilitates the diffusion of basic amino acids as well as some structurally analogous ß-lactam antibiotics like imipenem. Mutational inactivation of the oprD gene is associated with carbapenem resistance in Pseudomonas aeruginosa, whereas the C-terminal portion of OprD and, in particular, the hypothetical loop L7, was responsible for the unusual meropenem hypersusceptibility. There is also evidence that OprD porin of Pseudomonas aeruginosa bears protease activity with the catalytic activity established in residues His156, Asp208, and Ser296. However, not all family members have these residues conserved, suggesting that are not all enzymes. Various proteins of the family have shown different specificities in the uptake of metabolites including glycine-glutamate, histidine, proline, tyrosine, cis-aconitate and pyroglutamate. X-ray crystal structure of OprD from Pseudomonas aeruginosa has revealed a monomeric 18-stranded ß-barrel. | ||||
Representative image: 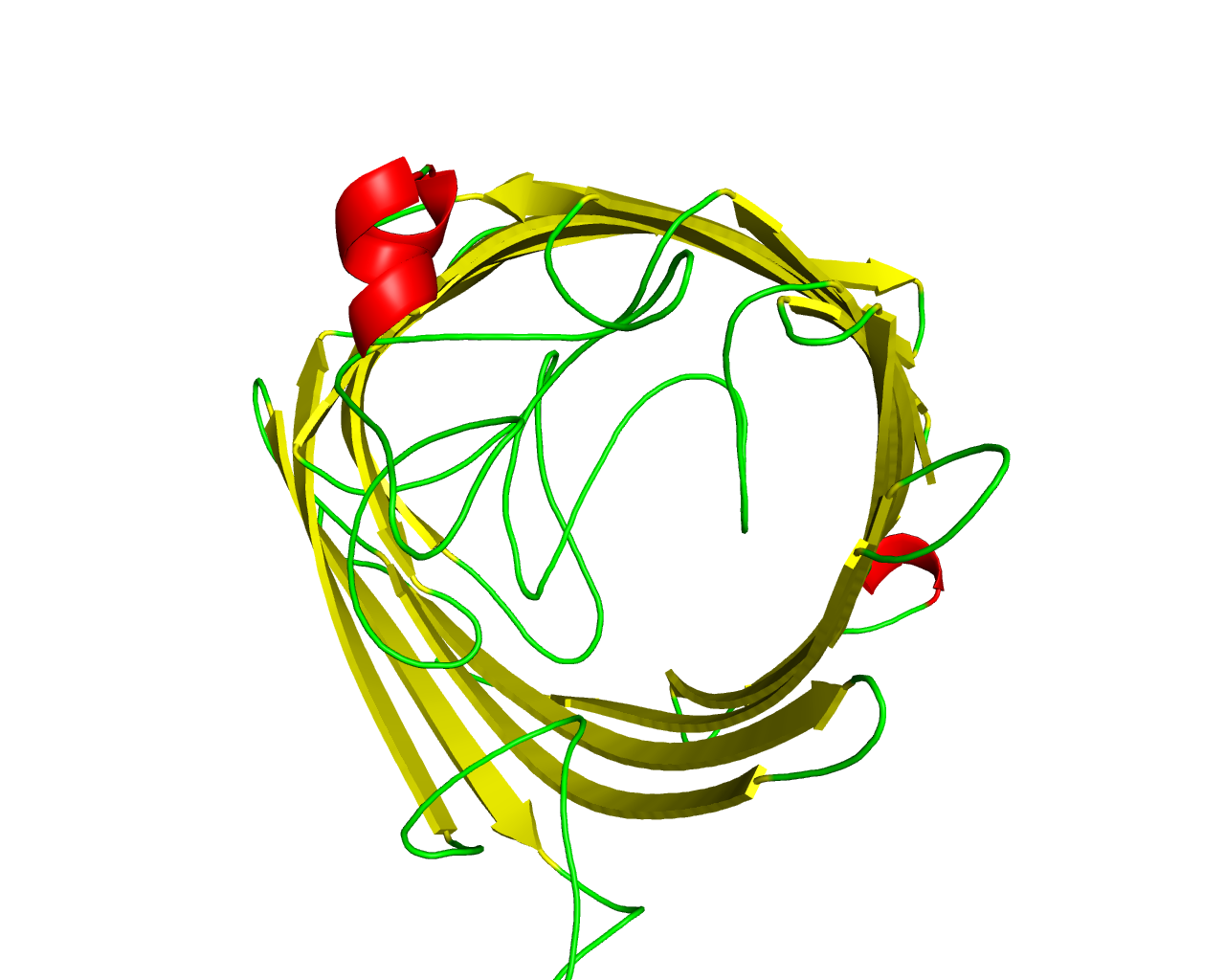 | ||||
| Literature references | ||||
Getting Drugs through Small Pores: Exploiting the Porins Pathway in Pseudomonas aeruginosa ACS Infect Dis. 2018 Oct 12;4(10):1519-1528. doi: 10.1021/acsinfecdis.8b00149. Epub 2018 Aug 8. PMID: 30039960 | ||||
Structure, function and regulation of Pseudomonas aeruginosa porins FEMS Microbiol Rev. 2017 Sep 1;41(5):698-722. doi: 10.1093/femsre/fux020. PMID: 28981745 | ||||
Structural Insights into Outer Membrane Permeability of Acinetobacter baumannii Structure. 2016 Feb 2;24(2):221-31. doi: 10.1016/j.str.2015.12.009. Epub 2016 Jan 21. PMID: 26805524 | ||||
Proteins causing membrane fouling in membrane bioreactors Water Sci Technol. 2015;72(6):844-9. doi: 10.2166/wst.2015.282. PMID: 26360742 | ||||
Substrate specificity within a family of outer membrane carboxylate channels PLoS Biol. 2012 Jan;10(1):e1001242. doi: 10.1371/journal.pbio.1001242. Epub 2012 Jan 17. PMID: 22272184 | ||||
Structure of a putative BenF-like porin from Pseudomonas fluorescens Pf-5 at 2.6 A resolution Proteins. 2010 Nov 1;78(14):3056-62. doi: 10.1002/prot.22829. PMID: 20737437 | ||||
Structural insight into OprD substrate specificity Nat Struct Mol Biol. 2007 Nov;14(11):1108-9. doi: 10.1038/nsmb1304. Epub 2007 Oct 21. PMID: 17952093 | ||||
Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa J Bacteriol. 2006 Jan;188(1):45-54. doi: 10.1128/JB.188.1.45-54.2006. PMID: 16352820 | ||||
Analysis of the Pseudomonas aeruginosa oprD gene from clinical and environmental isolates Environ Microbiol. 2002 Dec;4(12):872-82. doi: 10.1046/j.1462-2920.2002.00281.x. PMID: 12534469 | ||||
Function of pseudomonas porins in uptake and efflux Annu Rev Microbiol. 2002;56:17-38. doi: 10.1146/annurev.micro.56.012302.160310. Epub 2002 Jan 30. PMID: 12142471 | ||||
C-terminal region of Pseudomonas aeruginosa outer membrane porin OprD modulates susceptibility to meropenem Antimicrob Agents Chemother. 2001 Jun;45(6):1780-7. doi: 10.1128/AAC.45.6.1780-1787.2001. PMID: 11353625 | ||||
Amino acid-mediated induction of the basic amino acid-specific outer membrane porin OprD from Pseudomonas aeruginosa J Bacteriol. 1999 Sep;181(17):5426-32. doi: 10.1128/JB.181.17.5426-5432.1999. PMID: 10464217 | ||||
Identification of the catalytic triad of the protein D2 protease in Pseudomonas aeruginosa Biochem Biophys Res Commun. 1998 Jun 9;247(1):142-5. doi: 10.1006/bbrc.1998.8745. PMID: 9636669 | ||||
| Proteins in this family with 3D-structure | ||||
View Entry | Description | Organism | Length | # strands |
